Complex Networks & Systems
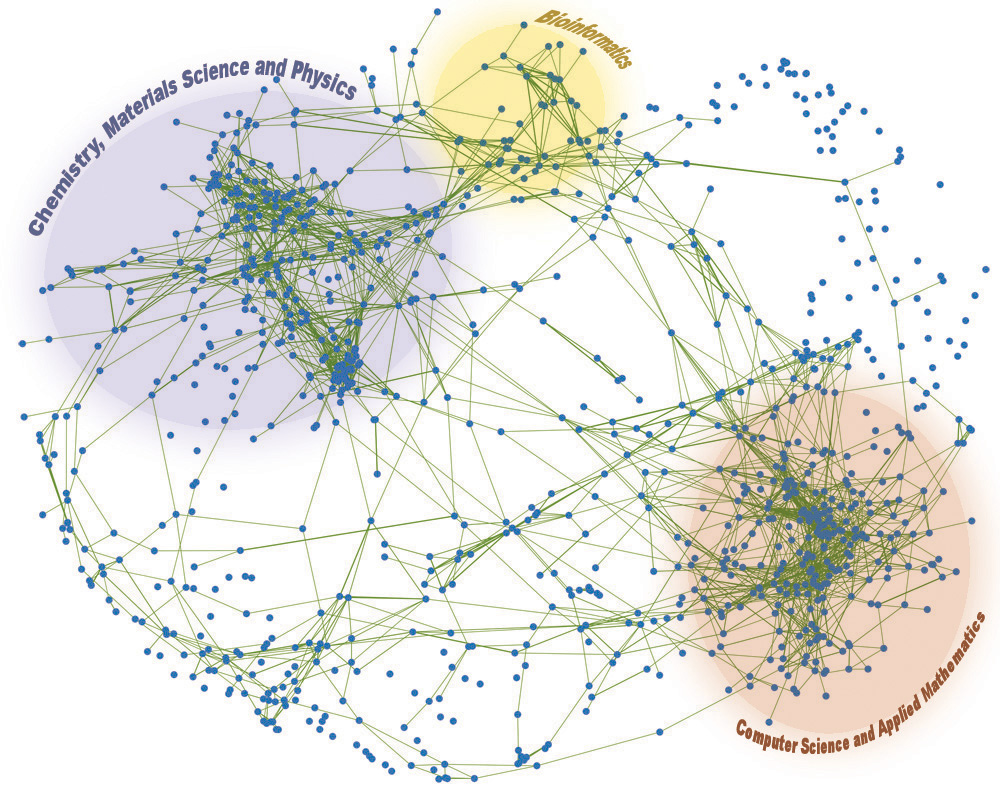
Networks are mathematical objects used to study multivariate systems. The most commonly used network representation is the graph – where multivariate dynamics is reduced to static interactions – but many other formalisms exist such as hyper-graphs, relations, and dynamical systems theory. With the availability of large-scale data about many domains ranging from molecular biology to social interactions, Network Science has made a substantial leap in providing predictive models of many complex problems – in addition to important contributions to theory with conceptual models. Most of these advances have been achieved using solely graph theory, without considering multivariate dynamics. In our CASCI group we are working on various projects which use both static and dynamical representations of networks to make inferences about multivariate complex systems.

Control and Redundancy in Network Dynamics
As the complex network field matures, there is a need to move beyond understanding network topology to controlling multivariate dynamics, or collective behavior. This is particularly true in systems biology and medicine, where increasingly accurate models of biochemical regulation have been produced. More than understanding the organization of biochemical regulation, we need to derive control strategies that allow us, for instance, to move a mutant cell to a wild-type state, or revert a mature cell to a pluripotent state. We have been developing methods to remove redundancy from automata rules to reveal their canalization properties, thus simplifying the characterization of control in large models of natural networks, such as systems biology models of biochemical regulation. Our studies have demonstrated that canalization (measured as effective connectivity) is an order parameter of Boolean Network (BN) dynamics, and a major factor in network controllability. We have also been working on methods to extract modularity and multi-scale integration in multivariate dynamical systems, such brain activity and biochemical regulation.
MoreRedundancy in Network Topology
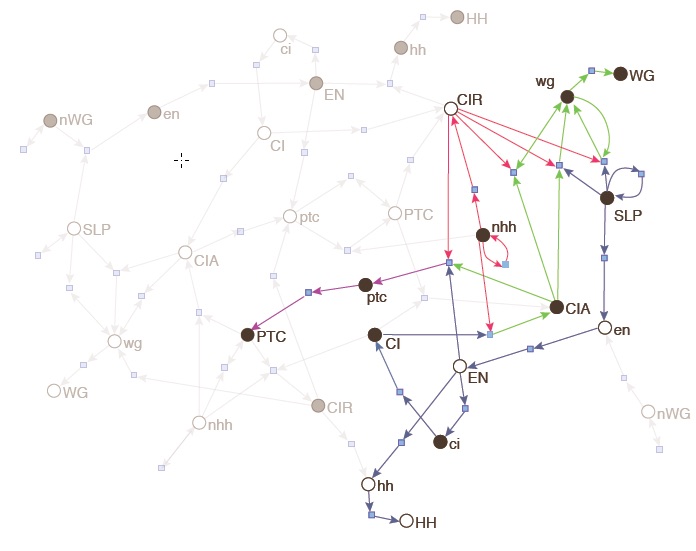
The structure of networks has provided many insights into the organization of complex systems. The success of this approach is its ability to capture the organization of multivariate interactions, and how it changes in time (network evolution) without explicit dynamical rules for node variables. When networks are modeled with weighted graphs, however, unexpected topological properties arise. For instance, distances between nodes tend to break the triangular inequality, leading to semi-metric topologies. We have been studying the causes and effects of such semi-metric topologies via an isomorphism between distance and proximity graphs. Our study has revealed that redundancy abounds in networks built from real-World data ranging from brain to social networks, meaning that most edges are not necessary to compute shortest-paths. This has led to new theoretical concepts such as distance closures and distance backbones, as well as to successful applications to problems such as recommender systems, relational inference of biochemical interactions, computational fact-checking, etc.
Knowledge Networks & Web Intelligence
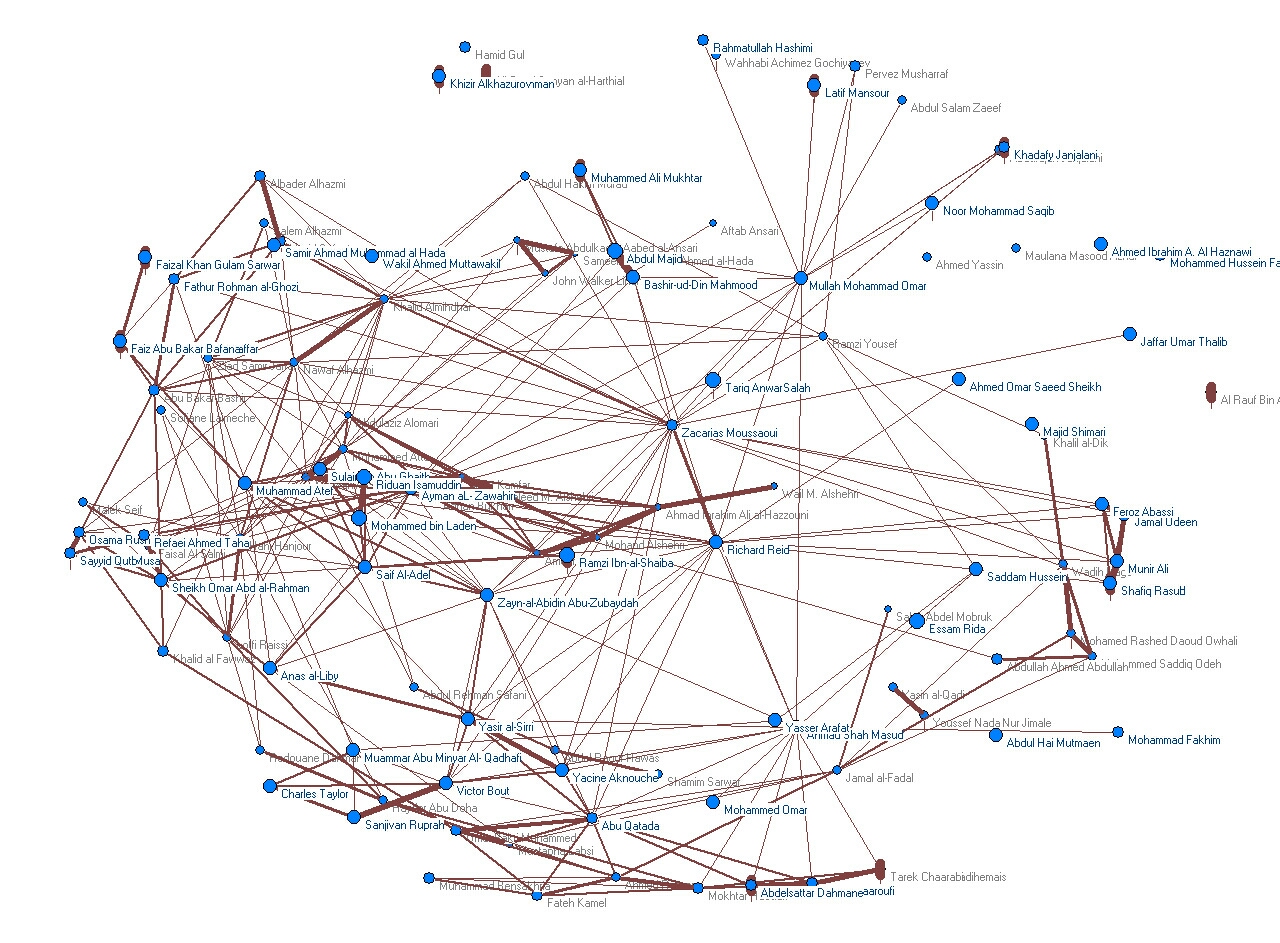
Complex networks can be built from multivariate data via a variety of measures (e.g. co-occurrence) to characterize the strength of variable interaction. We have used such networks to capture the collective knowledge of the communities of users of online resources, such as the scientific literature in PubMed and digital libraries (the “Bibliome”), Wikipedia, as well as social media such as Twitter and Instagram. We have shown that such simple but large-scale knowledge representations allow us to build competitive informatics technology for recommender systems, relational inference of biochemical interaction, and even computational fact-checking.
MoreAgent-Based Modeling
We have contributed to understanding complex adaptive systems by developing agent-based models of systems whose evolutionary role or adaptive capabilities are not well understood. For instance, we provided the first computational model that demonstrates the evolutionary potential of RNA Editing and a model that proves the ability of T-Cell cross-regulation to classify self from non-self in the presence of populations of changing pathogens. We have also developed bio-inspired methods for spam detection and (biomedical) text classification from that model. More generally, we have contributed to the study of the interplay between self-organization and natural selection by introducing the concept of selected self-organization and developing evolutionary algorithms and agent-based models to study it. We have been working on various other applications of the agent based modeling framework, such as evolving cellular automata and adaptive agents for recommendation systems.
More