Biomedical Literature and Social Media Mining
Our group has been involved in this field from its very start, having participated successfully in the first four BioCreAtIvE (Critical Assessment for Information Extraction) between 2004 and 2012. Much of the research presently conducted in the biomedical domain relies on the inference of correlations and interactions from data at multiple levels of the biological organization: from the molecular to the social. Because we ultimately want to increase our knowledge of the biochemical, functional and behavioral roles of genes and proteins in organisms, there is a clear need to integrate the associations and interactions among biological entities that have been reported and accumulated in experimental databases, the literature, electronic health records, and non-traditional data sources such as social media.
Biomedical literature mining is an important informatics methodology for large scale information extraction from repositories of textual documents, as well as for integrating information available in various domain-specific databases and ontologies, ultimately leading to knowledge discovery. It helps uncover relationships and interactions buried in the literature and other media, from experimental to phenomenological data. The goal is then to tap into the vast biomedical collective knowledge available in various data sources, which we can think of as the "bibliome" or the "digital phenotype" in health and disease.
Our contributions to the Biomedical literature mining have been the development of novel methods based on network science or bio-inspired computing. This data-driven approach has enabled the automatic discovery, classification and annotation of protein-protein and drug-drug interactions, health risks including gender and age biases, pharmacokinetic parameters in drug interaction and adverse reaction studies, population and epidemiological studies, protein sequence and structure prediction, functional annotation of transcription data, enzyme annotation publications, etc. Examples of these are shown below, together with links to additional resources and publications.
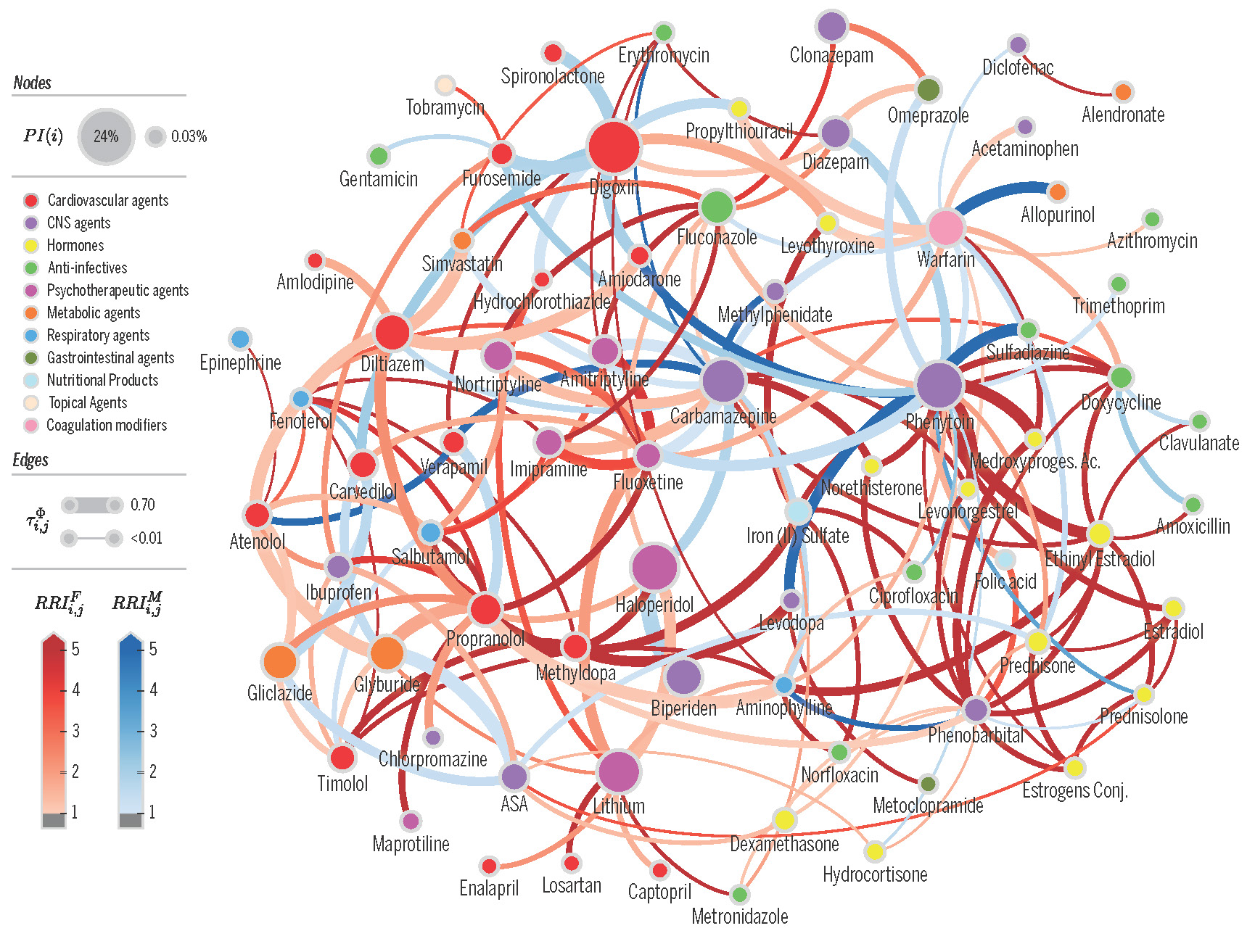
Proximity network of drug-drug interactions (DDI network) extracted from a electronic health records of a large population. Nodes denote drugs involved in at least one co-administration known to be a DDI. Node color represents the highest level of primary action class, as retrieved from Drugs.com (see legend). Node size represents the probability of interaction. Edge weights denote risk in population. Edge colors denote edges that are higher risk for females (blue) or males (red). From Correia et al [2019]
Public health monitoring using Social Media data
Social media and mobile application data enable population-level observation tools with the potential to speed translational research. Our group has been one of the first to use social media data to study collective social behavior in biomedical problems [Correia, Wood, Bollen & Rocha, 2020]---see also our related work in computational social science. For instance, our group was the first to use Instagram to build public health monitoring and surveillance tools for discovering drug interactions, adverse reactions, and behavior pathology, focusing on depression and epilepsy [Correia, Li, & Rocha, 2016]. This recent work demonstrates that the universe of social media provides a very promising source of large-scale data that can help with monitoring and understanding public health in ways that have not been hitherto possible. Indeed, given the large number of users, social media data allows us to identify under-reported, population-level pathology.
The Social Network of Healthcare - How Instagram and Twitter are Providing New Insights. Luis Rocha explains the new software-driven approach to medical research. Big data generated through social media such as Twitter and Instragram provides leads to actionable insights to improve the efficacy of prevention and treatment. [Correia, Wood, Bollen & Rocha, 2020]
Our methodology is based on the longitudinal analysis of social media user timelines at different timescales: day, week or month. Our approach enables extraction and study of social media cohorts with great demographic, geographic, and situational precision, which is important for social media mining in biomedical studies as these require attention to cohort segmentation. Most social mining approaches focus on collecting large sets of random tweets, selected by hashtags or other textual elements, which confound geographical and demographic cohorts. Instead, we focus on large-scale, longitudinal cohort datasets comprised of the entire timelines of users selected for having a tweet with a hashtag (e.g. a medication [Correia, Li & Rocha, 2016 ]). We have shown that such longitudinal social media datasets are better suited for building precise cohorts, as they provide enough data and resolution to predict adverse drug reactions and interactions, as well characterize subpopulations with specific health concerns (e.g. epilepsy) [Correia, Li & Rocha, 2016 ] and other problems of biomedical relevance [Correia, Wood, Bollen & Rocha, 2020].
Typically, our methodology involves building knowledge (weighted) graphs from the co-occurrence of terms from various biomedical dictionaries (drugs, symptoms, natural products, side-effects, and sentiment) at various timescales. We showed that spectral methods, shortest-paths, and distance closures reveal relevant drug-drug and drug-symptom pairs, as well as clusters of terms and drugs associated with the complex pathology associated with epilepsy and depression. We validate inferences about drug interactions and adverse reactions via curated bioinformatics databases (e.g. DrugBank and SIDER), and develop demo tools to share our analysis with the community [Correia, Li, & Rocha, 2016, Min et al, 2021]. We currently analyze various web search and social media sources such as: Google Trends, Wikipedia, Twitter, Facebook, ChaCha, Reddit, and the Epilepsy Foundation public forums, and have focused on studying depression, epilepsy, and opioid abuse, as well as other health-related problems such as human-reproduction and even automated online fact-checking. Very important for this area is also the development of biomedical corpora and dictionaries to mine social media and the literature, as well as analysis of electronic health records described below. See publications below for details on all these threads.
Studying Drug-Drug interaction from the Literature and Electronic Health Records
Drug-drug interactions (DDIs) are major causes of morbidity and mortality and a subject of intense scientific interest. Biomedical literature mining can aid DDI research by extracting evidence for large numbers of potential interactions from published literature and clinical databases. We started with the estimation of pharmacokinetics numerical data from literature to mine drug-specific (e.g. Midazolam (MDZ)) pharmokinetic (PK) clearance data (systemic and oral) from the literature. We obtained 88% precision rate and 92% recall rate are achieved, with an F-score = 90%. Out-performs support vector machine (F-score of 68.1%). Further investigation on 7 other drugs showed comparable performance [Wang et al, 2009]. Recently, via a four-year ($1.7M) R01 grant from from NIH/NLM we have studied the large-scale extraction of drug-Interaction from medical text. This is a collaboration with Prof. Lang Li from Ohio State Medical School, and Prof. Hagit Shatkay from the University of Delaware. While evidence for DDI ranges in scale from intracellular biochemistry to human populations, literature mining methods have not been used to extract specific types of experimental evidence which are reported differently for distinct experimental goals. We have developed and used the team's manually curated corpora [Wu et al, 2013; Zhang et al, 2022] of PubMed abstracts and annotated sentences with three types of experimental DDI evidence: in vitro, in vivo, and clinical. The goal is the production of a text mining pipeline using several linear classifiers and a variety of feature transformation methods. Preliminary results [Kolchinsky et al 2015] on pharmacokinetics DDI experimental evidence in PubMed has yielded excellent classification performance in distinguishing relevant and irrelevant abstracts (reaching F1 ~= 0.93, MCC ~= 0.74, iAUC ~= 0.99) and sentences (F1 ~= 0.76, MCC ~= 0.65, iAUC ~= 0.83). New results on all three DDI types are forthcoming.
Our group and collaborators have also studied the DDI phenomenon using other sources of large-scale data such social media [Correia, Wood, Bollen & Rocha, 2020; Correia, Li, & Rocha, 2016] (see above) and electronic health records(EHR) [Correia et al, 2019 ]. In addition to uncovering the most worrisome DDI prescribed to large populations, at great health and financial cost to individuals and communities, our analysis of EHR revealed very significant gender and age biases, whereby women and older people are prescribed many more DDIs than expected by random given the same rates of medication co-prescription. In forthcoming work with Prof. Alfonso Valencia's group, we extend this study to various worldwide populations and also study comorbidity in medical care.

A classification pipeline for DDI-relevant abstracts and evidence sentences. The pipeline includes selection of corpus documents, hand-labeling of ground truth assignments, extraction and normalization of textual features, and computation of unigram/bigram occurrences matrices. Cross-validation folds are used to estimate generalization performance of classifier and feature transform configurations, while nested (inner) cross-validation folds are used to choose classifier hyperparameters. From Kolchinsky et al [2015]

PPI task- Decision structure on the protein-protein interaction article test data of Biocreative II, as produced by our Variable Trigonometric Threshold model.Abi Haidar, A et al. (2008)
Protein-Protein Interaction Discovery (PPI)
We have worked in the discovery and automatic annotation of relationships among biochemical entities, e.g. protein-protein and gene-disease interactions. The Biocreative challenges II, II.5, and III, which we participated in [Abi-Haidar et al,2008], [Kolchinsky et al, 2010], [Lourenco et al, 2011]), included a series of tasks on extraction of protein-protein interaction information from the literature. As the field moves to uncovering relations rather than entities, our complex network approach to biomedical literature mining [Verspoor et al,2005], which we tried on the first BioCreative competition, makes all the more sense. Additionally, since literature mining hinges on the quality of available sources of literature as well as their linkage to other electronic sources of biological knowledge, it is particularly important to study the quality of the inferences it can provide. We were among most competitive teams in the PPI tasks of BioCreative II, II.5 and III. We have also developed a bio-inspired solution for binary classification of textual documents inspired by T-cell cross-regulation in the vertebrate adaptive immune system. See our publications below for additional details
Characterizing gene regulation
Spectral methods such as Singular Value Decomposition (SVD), are very useful for tasks ranging from gene expression analysis [Wall, Rechtsteiner and Rocha, 2003] to automatic functional annotation of genes and proteins from the literature [Rechtesteiner, 2005; Maguitman, A. et al, 2006; Haidar et al, 2008;]. We have studied SVD-based methods for visualization of gene expression data, representation of the data using a smaller number of variables, and detection of patterns in noisy gene expression data. SVD ("eigen-clustering") of microarray data produces sets of co-expressed genes, which were then characterized with annotations automatically extracted from literature .
More recently, we have used SVD and information theory to cluster very large knowledge networks of gene regulation obtained from bioinformatics databases and the literature. This allows us to identify overlapping functional clusters that occur in various scales of complex networks [Correia, Navarro-Costa and Rocha, 2020], such as those characterizing gene regulation. Together with our distance backbone methodology, this has lead to the discovery of novel genes involved in human infertility [Correia et al, 2022].

Rechtsteiner, A. (2005). PhD Dissertation.

PSP task - Our combined method performs significantly better than either the original structure predictionor keyword based prediction methods alone. Rechtsteiner, A., et al (2006)
Protein Structure Prediction (PSP)
Linking of information from different data sources, specifically literature, becomes increasingly important to annotate the growing number of new genome sequences. For the large percentage of genes with no known sequence homologs, new, possibly integrative, methods need to be developed. Ab-initio structure prediction and comparison is a method some of us pursued previously for functional annotation of sequences with no known homologs. We used a large set of sequences of known structure to evaluate a literature-based method against previously used ab-initio structure prediction methods. The Literature-mining prediction is comparable to best ab-initio methods in lack of sequence homology. Combining text-mining with ab-initio method leads to 35% improvement over ab-initio method alone. See [Rechtsteiner et al, 2006]
Protein Family Prediction (PFP)
Since literature mining hinges on the quality of available sources of literature as well as their linkage to other electronic sources of biological knowledge, it is particularly important to study the quality of the inferences it can provide. We have been working in the large-scale validation of bibliome algorithms , and proposed a method that predict a protein’s Pfam family correctly 76% of the time and 89% of the time issue a prediction that will be among top 5 families [Maguitman et al,2006].

Proteins voting in proportion to their cosine similarity to the target protein. Maguitman, A. et al (2006)
Funding Project partially funded by
- myAURA: Personalized Web Service for Epilepsy Management. National Institutes of Health, National Library of Medicine Program, 1R01LM012832-01, October 2018-2022
- Fundacao para a Ciencia e Tecnologia, Portugal. DSAIPA/AI/0087/2018. Project title: “Identification and Forecasting Hospital Emergency Demand”. 2018-2021
- National Science Foundation, Research Traineeship Program, NSF1735095: Interdisciplinary Training in Complex Networks and Systems, 2017-2022
- NATIONAL INSTITUTES OF HEALTH, NATIONAL LIBRARY OF MEDICINE PROGRAM, OCTOBER 2014/SEPTEMBER 2019. Project Title: R01LM011945-01 BLR: Evidence-based Drug-Interaction Discovery: In-Vivo, In-Vitro and Clinical.
- Fulbright U.S. Scholar grant, J. William Fulbright Foreign Scholarship Board (FFSB). 2016-2017
- Precision Health Initiative, "Population Health and Data and Informatics Clusters, Indiana Cohort Enhancement Study (4K Hoosiers)", Indiana University, 2017-2019
- Fundação Luso-Americana para o Desenvolvimento (Portugal), Program with National Science Foundation (USA), Proj 276/2016, 2017-2018. "Large-scale analysis of social network data for detecting drug-interaction via population behavior
- Complex Systems and Health Project Development Team, Indiana Clinical Translational Sciences Institute (ICTSI) NIH/NCRR UL1TR001108. "Sudden Unexpected Death in Epilepsy: Identifying Risk Factors with Social Media Mining" . 2016-2018
- PERSISTENT SYSTEMS, INC., 2014-2017. Project Title: Large-Scale Text and Social Data Analytics for Health.
- PRECISION HEALTH INITIATIVE, INDIANA UNIVERSITY, 2017-2019. Population Health and Data and Informatics Clusters, Indiana Cohort Enhancement Study (4K Hoosiers).
- FUNDAÇÃO LUSO-AMERICANA PARA O DESENVOLVIMENTO (PORTUGAL) AND NATIONAL SCIENCE FOUNDATION (USA), 2016-2018. Project Title: Large-scale analysis of social network data for detecting drug-interaction via population behavior
- INDIANA CLINICAL TRANSLATIONAL SCIENCES INSTITUTE (ICTSI), COMPLEX SYSTEMS and HEALTH PROJECT DEVELOPMENT TEAM, NIH/NCRR UL1TR001108. Aug. 2016/ Feb. 2018. Project Title: Sudden Unexpected Death in Epilepsy: Identifying Risk Factors with Social Media Mining.
- Indiana University Collaborative Research Grants 2011. Project title: “Drug-Drug Interaction Prediction from Large-scale Mining of Literature and Patient Records”.
- Fundação Luso-Americana para o Desenvolvimento (Portugal) and National Science Foundation (USA), 2012-2014. Project title: “Network Mining For Gene Regulation And Biochemical Signaling.” (171/11)
Project Members (Current and Former)

Luis Rocha (PI)

Alaa Abi-Haidar

Rion Brattig Correia

Joana Gonçalves-Sá

Jack Felag

Ziqi Guo

Lang Li

Jasleen Kaur

Artemy Kolchinsky

Analia Lourenco

Ana Maguitman

Wendy Miller

Aehong Min

Paulo Navarro Costa

Thomas Parmer

Predrag Radivojac

Andreas Rechtsteiner

Jordan C. Rozum

Jon Sánchez-Valle

Hagit Shatkay

Tiago Simas

Alfonso Valencia

Xuan Wang

Zhiping (Paul) Wang
Selected Project Publications
- R.B. Correia, J.C. Rozum, L. Cross, J.Felag, M. Gallant, Z. Guo, B.W. Herr II, A. Min, J. Sanchez-Valle, D.S. Rocha, A. Valencia, X. Wang, and K.Börner, W.R.Miller, and L.M. Rocha. [2025]. "myAURA: Personalized health library for epilepsy management via knowledge graph sparsification and visualization". Journal of the American Medical Informatics Association, ocaf012. DOI: 10.1093/jamia/ocaf012. Preprint also available: arXiv:2405.05229.
- J. Sánchez-Valle, M. Flores-Rodero, F.X. Costa, J. Carbonell-Caballero, I. Núñez-Carpintero, R. Tabarés-Seisdedos, L.M. Rocha, D. Cirillo, A. Valencia [2025]. "Sex-specific transcriptome similarity networks elucidate comorbidity relationships." bioArXiv Preprint. DOI: 10.1101/2025.01.22.634077. Preprint also available: arXiv:2405.09649.
- Z. Guo, J. Felag, J.C. Rozum, R.B. Correia, and L.M. Rocha. [2024]. "Selecting focused digital cohorts from social media using the metric backbone of biomedical knowledge graphs.". arXiv:2405.07072. DOI: 10.48550/arXiv.2405.07072.
- A. Min, X. Wang, R.B. Correia, J. Rozum, W.R. Miller, and L.M. Rocha. [2024]. "Refinement of an Epilepsy Dictionary through Human Annotation of Health-related posts on Instagram.". arXiv:2405.08784. DOI: 10.48550/arXiv.2405.08784.
- J. Sánchez-Valle, R.B. Correia, M. Camacho-Artacho, R. Lepore, M.M. Mattos, L.M. Rocha, A. Valencia [2024]. "Prevalence and differences in the co-administration of drugs known to interact: an analysis of three distinct and large populations.". BMC Medicine 22: 166. doi: 10.1186/s12916-024-03384-1. PMID: 38637816.
- R.B. Correia, J.M. Almeida, Ma.J. Wyrwoll, I.Julca, D. Sobral, C.S. Misra, L.G. Guilgur, H.-C. Schuppe, N. Silva, P. Prudêncio, A. Nóvoa, A. S. Leocádio, J. Bom, M. Mallo, S. Kliesch, M. Mutwil, L.M. Rocha, F.Tüttelmann, J. D. Becker, P. Navarro-Costa [2024]. "The conserved genetic program of male germ cells uncovers ancient regulators of human spermatogenesis". eLife.Reviewed reprint: eLife-RP-TR-2024-95774. In Press. bioRxiv 2022.03.02.482557, doi: 10.1101/2022.03.02.482557.
- A. Min, W.R. Miller, L.M. Rocha, K. Borner, R.B. Correia, and P.C. Shih. [2023]. "Understanding Contexts and Challenges of Information Management for Epilepsy Care". In: Proceedings of the 2023 CHI Conference on Human Factors in Computing Systems (CHI ’23), April 23–28, 2023, Hamburg, Germany. ACM, New York, NY, USA, 15 pages. DOI:/10.1145/3544548.3580949. Preprint Available.
- S. Zhang, H. Wu, L. Wang, G. Zhang, L.M. Rocha, H. Shatkay, L. Li[2022]. "Translational drug–interaction corpus". Database, baac031. doi: 10.1093/database/baac031.
- I.B. Wood, R.B. Correia, W.R. Miller, and L.M. Rocha [2022]. "Small Cohort of Epilepsy Patients Showed Increased Activity on Facebook before Sudden Unexpected Death". Epilepsy & Behavior. 128: 108580. DOI:10.1016/j.yebeh.2022.108580. Preprint: arXiv:2201.07552.
- A. Min, W. Miller, L.M. Rocha, K.Börner, R.B. Correia, and P.C. Shih. [2021]. "Just In Time: Challenges and Opportunities of First Aid Care Information Sharing for Supporting Epileptic Seizure Response".Proceedings of the ACM on Human-Computer Interaction.. 5, CSCW1, Article 113, pp. 1-24. DOI:10.1145/3449187.
- R.B. Correia, I.B Wood, J. Bollen, L.M. Rocha [2020]. "Mining social media data for biomedical signals and health-related behavior". Annual Review of Biomedical Data Science . 3(1). DOI: 10.1146/annurev-biodatasci-030320-040844.
- Correia, R.B., P. Navarro-Costa, and L.M. Rocha [2020]. "Extraction of overlapping modules in networks via spectral methods and information theory". Complex Networks 2020. The 9th International Workshop on Complex Networks and Their Applications. Dec. 1-3, 2020, Madrid, Spain (Online).
- H.-Y. Wu, A. Shendre, S. Zhang, P. Zhang, L. Wang, D. Zeruesenay, L.M. Rocha, H. Shatkay, S. Quinney, X. Ning, and L. Li [2019]. Translational Knowledge Discovery between Drug Interactions and Pharmacogenetics. Clinical Pharmacology & Therapeutics . 2019 Dec 21. doi: 10.1002/cpt.1745
- R.B. Correia, L.P. de Araújo, M.M. Mattos, L.M. Rocha [2019]. City-wide Analysis of Electronic Health Records Reveals Gender and Age Biases in the Administration of Known Drug-Drug Interactions. NPJ Digital Medicine. 2:74
- M. ten Thij, I.B. Wood, L.M. Rocha, J. Bollen [2019]. "Detecting eigenmoods in individual human emotions." 5th International Conference on Computational Social Science (IC2S2) July 17-20, 2019, University of Amsterdam, The Netherlands.
- A. Min, W.R. Miller, L.M. Rocha, K. Borner, R.B. Correia, P.C. Shih [2019]. "Understanding Health Information Management of People with Epilepsy and Their Caregivers." Workshop on Interactive Systems in Health Care, The ACM CHI Conference on Human Factors in Computing Systems. May 4-9, 2019, Glasgow, U.K.
- I.B Wood, P.L. Varela, J. Bollen, L.M. Rocha, J. Gonçalves-Sá [2017]. Human Sexual Cycles are Driven by Culture and Match Collective Moods. Scientific Reports. 7: 17973 DOI: 10.1038/s41598-017-18262-5.
- R.B. Correia, L. Li, L.M. Rocha [2016]. “Monitoring potential drug interactions and reactions via network analysis of Instagram user timeliness“. Pacific Symposium on Biocomputing. 21:492-503.
- A. Kolchinsky, A. Lourenço, H. Wu, L. Li, and L.M. Rocha.[2015] “Extraction of Pharmacokinetic Evidence of Drug-drug Interactions from the literature.” PLoS ONE 10(5): e0122199. doi:10.1371/journal.pone.0122199.
- Wu, Hengyi, S. Karnik, A. Subhadarshini, Z. Wang, S. Philips, X. Han, C. Chiang, L. Liu, M. Boustani, L.M. Rocha, S.K. Quinney, D.A. Flockhart and L. Li [2013]. “An Integrated Pharmacokinetics Ontology and Corpus for Text Mining”. BMC Bioinformatics. BMC Bioinformatics. 14:35. DOI:10.1186/1471-2105-14-35. (Highly Accessed)
- A. Kolchinsky, A. Lourenço, L. Li, L.M. Rocha [2013]. “Evaluation of linear classifiers on articles containing pharmacokinetic evidence of drug-drug interactions“. Pacific Symposium on Biocomputing, 2013. 18:409-420.
- Zhiping Wang [2012]. “Biomedical Literature Mining for Pharmacokinetics Numerical Parameter Collection.” Ph.D. Dissertation (Computer Science Program), Indiana University.
- A. Lourenço, M. Conover, A. Wong, A. Nematzadeh, F. Pan, H. Shatkay, and L.M. Rocha [2012].“Correction: A linear classifier based on entity recognition tools and a statistical approach to method extraction in the protein-protein interaction literature”. BMC Bioinformatics, 13: 180.
- S. Karnik, A. Subhadarshini, Z. Wang, L.M. Rocha, and L. Li [2011].”Extraction of drug-drug interactions using all paths graph kernel.” (pdf). In: Proceedings of the 1st Challenge task on Drug-Drug Interaction Extraction. I. Segura-Bedmar, P. Martínez, D. Sánchez. September, 7th, 2011, Huelva, Spain, pp. 83-88.
- A. Lourenço, M. Conover, A. Wong, A. Nematzadeh, F. Pan, H. Shatkay, and L.M. Rocha [2011].”A Linear Classifier Based on Entity Recognition Tools and a Statistical Approach to Method Extraction in the Protein-Protein Interaction Literature“. BMC Bioinformatics. 12(Suppl 8):S12
- M. Krallinger, et al [2011].”The Protein-Protein Interaction tasks of BioCreative III: classification/ranking of articles and linking bio-ontology concepts to full text“. BMC Bioinformatics. 12(Suppl 8):S3.
- A. Lourenço, M. Conover, A. Wong, F. Pan, Alaa Abi-Haidar, A. Nematzadeh, H. Shatkay, and L.M. Rocha [2010].”Testing Extensive Use of NER tools in Article Classification and a Statistical Approach for Method Interaction Extraction in the Protein-Protein Interaction Literature” (pdf). Proceedings of the BioCreative III Workshop 2010, Bethesda, Maryland, September 13-15, 2010.
- A. Kolchinsky, A. Abi-Haidar, J. Kaur, A.A. Hamed and L.M. Rocha [2010]. “Classification of protein-protein interaction full-text documents using text and citation network features.” IEEE/ACM Transactions On Computational Biology And Bioinformatics, 7(3):400-411. DOI: doi.ieeecomputersociety.org/10.1109/TCBB.2010.55
- Z. Wang, S. Kim, S.K. Quinney, Y. Guo, S.D. Hall, L.M. Rocha, and L. Li [2009]. “Literature mining on pharmacokinetics numerical data: A feasibility study“. Journal of Biomedical Informatics. 42 (4): 726-735.
- A. Lourenço; R.C. Carreira; D. Glez-Peña; J.R. Méndez; S.A. Carneiro; L.M. Rocha; F. Díaz; E.C. Ferreira; I.P. Rocha; F. Fdez-Riverola; M. Rocha [2009]. “BioDR: Semantic Indexing Networks for Biomedical Document Retrieval.” Expert Systems with Applications, 37, 3444–3453.
- A. Abi-Haidar, J. Kaur, A. Maguitman, P. Radivojac, A. Retchsteiner, K. Verspoor, Z. Wang, and L.M. Rocha [2008]. Uncovering protein interaction in abstracts and text using a novel linear model and word proximity networks“. Genome Biology. 9(Suppl 2):S11
- Maguitman, A. G., Rechtsteiner, A., Verspoor, K., Strauss, C.E., Rocha, L.M. [2006]. “Large-Scale Testing Of Bibliome Informatics Using Pfam Protein Families“. In: Pacific Symposium on Biocomputing11:76-87.
- Rechtsteiner, A., Luinstra, J., Rocha, L.M., Strauss, C.E., [2006]. “Use of Text Mining for Protein Structure Prediction and Functional Annotation in Lack of Sequence Homology“. In: Joint BioLINK and Bio-Ontologies Meeting 2006 (ISMB Special Interest Group).
- Verspoor, K., J. Cohn, C. Joslyn, S. Mniszewski, A. Rechtsteiner, L.M. Rocha, T. Simas [2005]. “Protein Annotation as Term Categorization in the Gene Ontology using Word Proximity Networks“. BMC Bioinformatics, 6(Suppl 1):S20. doi:10.1186/1471-2105-6-S1-S20
- Andreas Rechtesteiner [2005]. Multivariate Analysis of Gene Expression Data and Functional Information: Automated Methods for Functional Genomics . PhD Dissertation, Systems Science Program, Portland State University.
- Wall, Michael E., Andreas Rechtesteiner, and Luis M. Rocha [2003]. “Singular Value Decomposition and Principal Component Analysis “. In: A Practical Approach to Microarray Data Analysis. D. P. Berrar, W. Dubitzky, and M. Granzow (Eds.). Kluwer Academic Publishers, pp. 91-109.