Models of Genotype Editing
Models of Genotype and RNA Editing

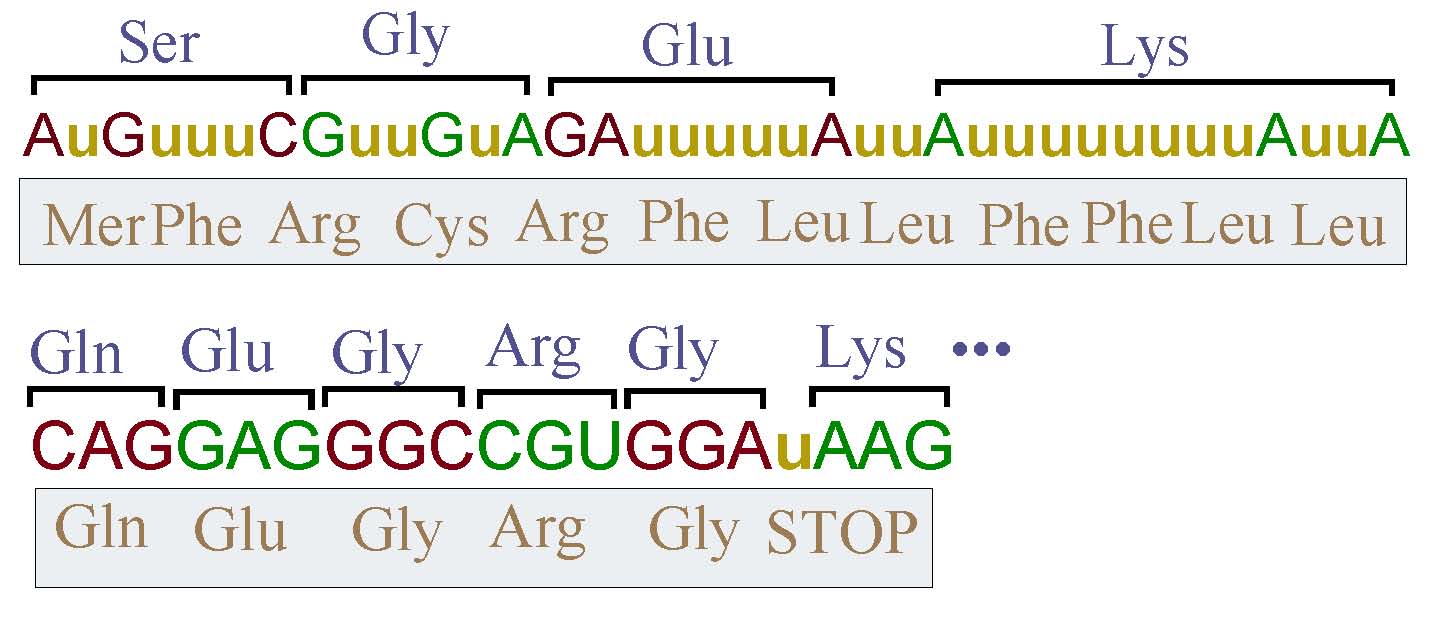
The discovery of messenger RNA (mRNA) molecules containing information not coded in DNA, first persuaded researchers in molecular biology that some mechanism in the cell might be responsible for post-transcriptional alteration of genetic information; this mechanism was called 'RNA Editing'. The term is used to identify any mechanism which will produce mRNA molecules with information not specifically encoded in DNA. Initially, the term referred to the insertion or deletion of particular bases (e.g. uridine), or some sort of base conversion (e.g. adenosine to guanisine). Today, many more RNA editing mechanisms, have been observed. The figure below describes two examples of uridine insertion in T-Brucei. The first example shows massive uridine insertion which changes dramatically the encoded peptide chain. The second example shows a single uridine insertion which provides a stop codon missing in the encoded gene.
In spite of the diversity and (presumed) differences in mechanism, research on different forms of RNA editing addresses the same questions: (1) what are the cis-acting RNA elements that designate a certain site for sequence alteration, (2) what are the trans-acting factors that operate in editing reactions and what is their mechanism of action, and (3) why do RNA editing processes exist?". Our research goal in this project is an investigation of the third question from a complex, evolutionary systems perspective. In particular we aim at:
- Understanding the theoretical nature of genetic systems with genotype editing, drawing from concepts developed in the fields of complex systems, artificial life, theoretical biology, and bio-semiotics.
- Developing computer simulations of populations of artificial organisms with different genotype editing capabilities, in order to investigate the hypotheses that genotype editing provides an evolutionary advantage to some organisms.
- Improve the state of the art on evolutionary algorithms
From our understanding of the RNA Editing System, we have extended the traditional Genetic Algorithm using several genetic editing characteristics that are gleaned from the RNA editing system as observed in several organisms. We have shown that the incorporation of editing mechanisms provides a means for artificial agents with genetic descriptions to gain greater phenotypic plasticity in dynamic environments, which may be environmentally regulated. In our more recent models, we study the evolution of agents with an artificial genome that contains both coding and non-coding components. The coding component encodes solutions to a particular fitness function or environment, while the non-coding component defines a set of editors which act on the coding component. We refer to the coding portion of the artificial genome as the codome, and to the non-coding portion as the editome. In each generation, the coding component of an agentís genotype, the codotype may be stochastically edited by the agentís non-coding editors, the editype, and produce a solution/phenotype different from what is encoded. Our simulations present novel insights as to how genotype editing allows agents to search fitness spaces differently from agents with non-edited genotypes. Edition is particularly useful in dynamic environments when the fitness function changes drastically. We are also investigating evidence to support the claim that genotype editing allows a wider exploration of the fitness space without sacrificing exploitation of good areas of the genotype space.
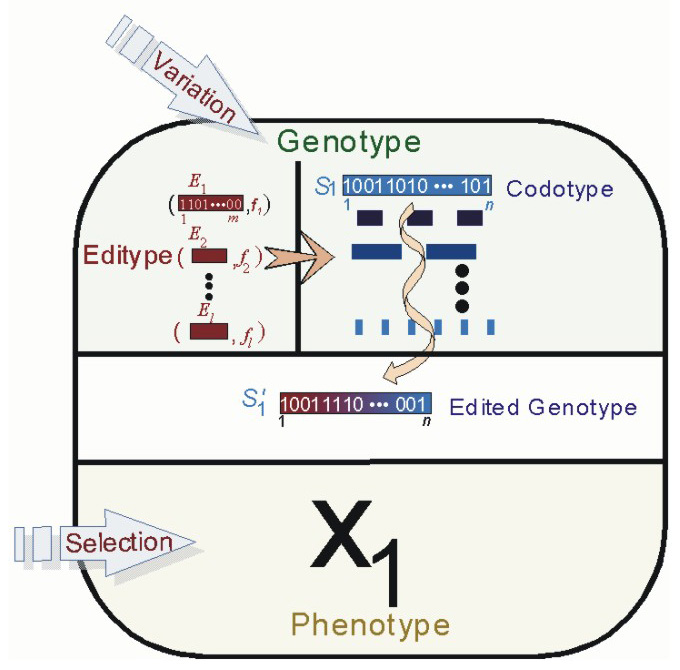
Agent with separate codotype and editype components of their genotype in our Evolutionary Model of Genotype Editing.
Publications on Genotype Editing

L.M. Rocha and J. Kaur [2007]."Genotype Editing and the Evolution of Regulation and Memory". Proceedings of the 9th European Conference on Artificial Life. Lecture Notes in Artificial Intelligence (LNAI), 4648: 63-73 (Springer-Verlag).
C. Huang, J. Kaur, A. Maguitman, L.M. Rocha[2007]."Agent-Based Model of Genotype Editing". Evolutionary Computation, 15(3): 253-89.
Rocha, L.M., A. Maguitman, C. Huang, J. Kaur, and S. Narayanan. [2006].""An Evolutionary Model of Genotype Editing". In: Artificial Life 10: Tenth International Conference on the Simulation and Synthesis of Living SystemsL.M.Rocha, L. Yaeger, M. Bedau, D. Floreano, R. Goldstone, and A. Vespignani (Eds.). MIT Press, In Press.
Huang, Chien-Feng and Luis M. Rocha [2005]. "Tracking Extrema in Dynamic Environments using a Coevolutionary Agent-based Model of Genotype Edition". In: Genetic and Evolutionary Computation Conference: GECCO 2005. ACM Press, pp. 545-552.
Rocha, Luis M. and Chien-feng Huang [2004]. "The Role of RNA Editing in Dynamic Environments". In: Ninth International Conference on the Simulation and Synthesis of Living Systems (ALIFE9). MIT Press, pp. 489-494
Huang, Chien-Feng and Luis M. Rocha [2004]. "A Systematic Study of Genetic Algorithms with Genotype Editing". In: Genetic and Evolutionary Computation: GECCO 2004. Lecture Notes in Computer Science Vol. 3102, pp.1233 - 1245. Springer-Verlag.
Huang, Chien-Feng and Luis M. Rocha [2003]. "Exploration of RNA Editing and Design of Robust Genetic Algorithms". 2003 IEEE Congress on Evolutionary Computation (CEC), Canberra, Australia, December 2003. R.Sarker et al (Eds). IEEE Press, pp. 2799-2806.
Rocha, Luis M. [2000]. "Syntactic autonomy, cellular automata, and RNA editing: or why self-organization needs symbols to evolve and how it might evolve them". In: Closure: Emergent Organizations and Their Dynamics. Chandler J.L.R. and G, Van de Vijver (Eds.) Annals of the New York Academy of Sciences. Vol. 901, pp 207-223.
Rocha, Luis M. [1997]." Evidence Sets and Contextual Genetic Algorithms: Exploring Uncertainty, Context, and Embodiment in Cognitive and Biological Systems. PhD Dissertation. State University of New York at Binghamton.
Rocha, Luis M. [1995]." Contextual Genetic Algorithms: Evolving Developmental Rules ." In: Advances in Artificial Life . F. Moran, A.Moreno, J.J. Merelo, and P. Chacon (Eds.). Series: Lecture Notes in Artificial Intelligence, Springer-Verlag. pp. 368-382.
Supplemental Materials for Publications

Oscillatory Small Royal Road Simulations
- Oscillation period 50: 4000 generations GA against ABMGE, first 1000 generations GA against ABMGE, Last 1000 generations GA against ABMGE, 4000 generations GA alone, 4000 generations ABMGE alone, and 4000 generations ABMGE with editype crossover alone
- Oscillation period 100: 4000 generations GA against ABMGE, first 1000 generations GA against ABMGE, Last 1000 generations GA against ABMGE, 4000 generations GA alone, 4000 generations ABMGE alone, and 4000 generations ABMGE with editype crossover alone
- Oscillation period 200: 4000 generations GA against ABMGE, first 1000 generations GA against ABMGE, and Last 1000 generations GA against ABMGE
Movies of Fitness Histograms
- Dynamic Schwefel Function
- Single run of GA (dynamic severity 50, 1000 generations)
- 10 runs of GA (dynamic severity 50, 1000 generations)
- Single run of ABMGE (dynamic severity 50, 1000 generations)
- 10 runs of ABMGE (dynamic severity 50, 1000 generations)
- Single run of ABMGE with editype crossover (dynamic severity 50, 1000 generations)
- 10 runs of ABMGE with editype crossover(dynamic severity 50, 1000 generations).


Averaged fitness of a population of agents in 10 runs of regular GA (top) and ABMGE (bottom) on the dynamic Schwefel Function (dynamic severity 50, 1000 generations) when the fitness function changes every 100 generations (shown in the movies as a yellow Background).